# Hot Search #
Scientists have been searching for methods to precisely control gene expression, which is crucial for fields, e.g., biosynthesis and gene cell therapy. A recent study has discovered that phytochrome accessory proteins derived from plants can play a significant role in mammalian cells.
On June 8, 2024, ECNU research team led by Ye Haifeng from the School of Life Sciences, the Shanghai Key Laboratory of Regulatory Biology, and the Center for Medical Synthetic Biology published a paper in Nature Communications titled “Exploring plant-derived phytochrome chaperone proteins for light-switchable transcriptional regulation in mammals”. This research marks the first discovery that plant-derived phytochrome chaperone proteins possess transcriptional activator activity in mammalian cells. Furthermore, they developed a novel light-switchable transcriptional regulation system (PTRC), opening up new possibilities for basic research in life sciences and biomedical engineering applications.

https://www.nature.com/articles/s41467-024-49254-5
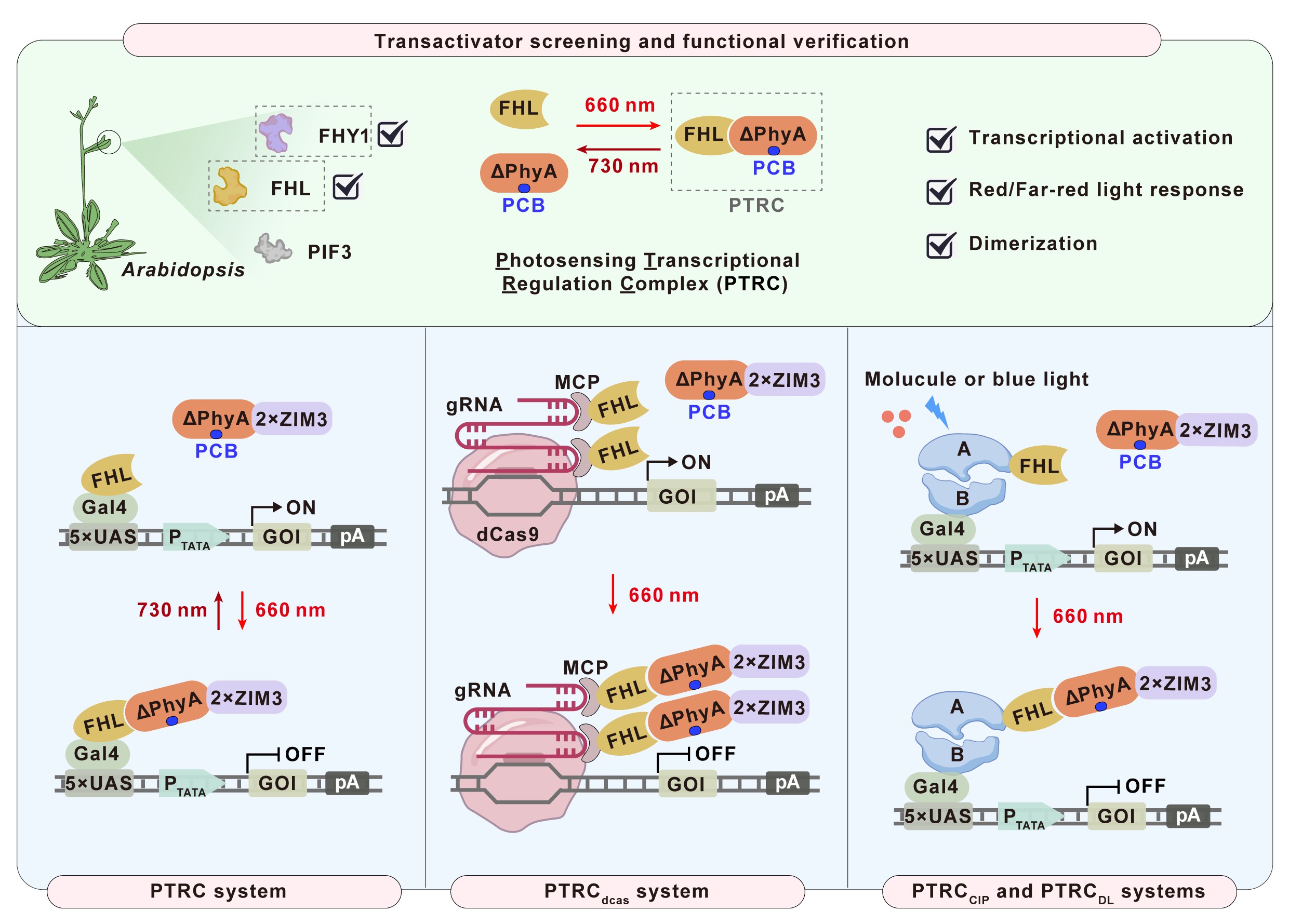
The birth of the PTRC system originated from a “failed experiment”. Back in 2019, when designing and developing the REDMAP system, researchers found that the expression of the reporter gene remained almost unchanged regardless of light or darkness, staying in a continuously activated state. They initially believed that PhyA and FHY1 might bind in the dark. However, upon further analysis, they discovered that the combinations capable of continuous activation in the dark were all fusion expressions of FHY1 with DNA-binding domains. Therefore, researchers boldly hypothesized that the plant-derived protein FHY1 might have transcriptional activator activity in mammalian cells.
To test this hypothesis, the research team fused FHY1 with different DNA-binding domains and validated the transcriptional activator functions of the plant-derived phytochrome chaperone proteins FHY1 and FHL from Arabidopsis in mammalian cells and mice. By leveraging their ability to heterodimerize with PhyA under light conditions, they constructed a light-switchable transcriptional regulation complex (PTRC). Under red and far-red light exposure, PTRC can switch between active and inactive states as easily as flipping a light switch.
To validate the effectiveness of this photosensitive system, researchers combined it with a gene-editing tool known as CRISPRa, developing a new platform capable of precisely controlling gene expression. Using this platform, scientists can regulate gene expression with light, enabling more flexible and accurate gene control.
Moreover, the research team integrated the PTRC system with other gene expression regulation modules, such as chemical small molecule or blue light-induction modules, creating various highly controllable systems. These systems not only performed excellently in laboratory settings but also demonstrated rapid and reversible gene regulation capabilities in live animal experiments.
This innovative research brings new hope to synthetic biology and biomedical engineering. By introducing plant-derived phytochrome chaperone proteins, scientists have developed a powerful and versatile tool, laying the foundation for future advanced research and medical applications. This breakthrough can be likened to plugging an intelligent switch into gene expression, making it more flexible and efficient.
Kong Deqiang, a PhD student of the Class 2020 at the School of Life Sciences, ECNU; Zhou Yang, a postdoctoral researcher at Wuhu Hospital Affiliated to East China Normal University; and Wei Yu, a graduate student of the Class 2021 at the School of Life Sciences, East China Normal University, are the co-first authors of the paper, with Researcher Ye Haifeng and Associate Researcher Guan Ningzi as the co-correspondents. The research was supported by the “Synthetic Biology” Key Special Project under the National Key R&D Program, the National Natural Science Foundation of China, and the Shanghai Science and Technology Commission Major Project on Synthetic Biology.
Source: ECNU School of Life Sciences
Copy editor: Philip Nash
Editor: Wicky Xu